SpeCollate: Deep Cross-Modal Similarity Network for Peptide Database Search using Mass Spectrometry Data
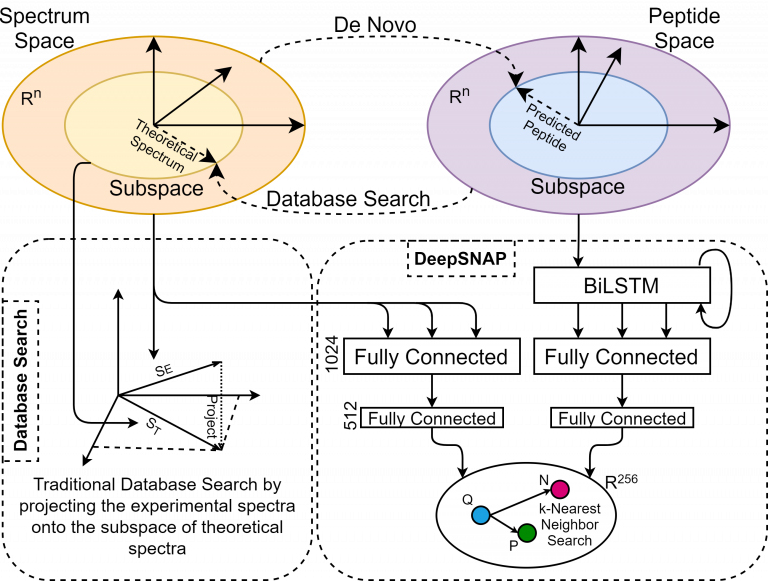
Historically, there have been two contrasting approaches for inferring peptides from mass-spectrometry data, i.e., de novo sequencing and database searching. The de novo approach tries to transform spectral space into peptide space by predicting individual amino acids from a given spectrum. On the other hand, database search tries to associate the experimental spectra to existing peptides by transforming peptide space into the spectral space and performing the comparisons. Each approach uses a heuristic similarity-scoring function to determine the match quality between an experimental spectrum and its corresponding peptide. However, when using heuristics, there is no solid reasoning outlining why a function is chosen over the other or why a particular feature within a process has the given associated weight. On the other hand, theoretical spectra are usually generated using a simple form of a simulator, introducing another source of inaccuracy in the search process.
Here we will discuss the design and implementation of a deep learning model called SpeCollate, which overcomes these issues by directly learning the similarity function between experimental spectra and peptide strings. SpeCollate transforms spectral and peptidal spaces into a shared Euclidean subspace by learning embeddings for spectra and peptides. The L2 distance between two data points in the resultant space directly correlates with their similarity.
By training the network on nearly 4.8 million sextuplets, obtained from the NIST and MassIVE peptide libraries, SpeCollate can achieve a promising peptide identification accuracy of up to ~99%.
Deep Learning methods are the next step in Big Data Proteomics. The limitations and oversights of the existing numerical techniques, bounded performance of spectral simulators, unoptimized scoring heuristics, inflated search space, and the opportunities made available by huge data repositories with annotated spectra are some of the key motivators behind this paradigm shift. Currently, there is no single heuristic from database search techniques that can claim as the most accurate strategy. Substantial work has been carried out towards developing computational methods for identifying peptides using database search and de novo algorithms. However, peptide identification problems are well-known and prevalent, including but not limited to misidentifications or no identifications for peptides, statistical accuracy (FDR), and inconsistencies between different search engines. Comparison across literature indicates decreased average accuracy of de novo algorithms (<35%) relative to database search algorithms (30-80%). Lack of quality assessment benchmarks makes the accuracy exhibited from these database search tools highly dependent on the data, indicating that further formal investigation and evaluation is warranted. Two significant sources of heuristic errors introduced in the numerical database search algorithms are how the peptide deduction takes place, i.e., simulation of the spectra (from peptides) and the peptide spectrum match scoring-function. The simplistic and a priori nature of the scoring mechanism neglects the MS data (and the database) that are under consideration, leading to variable quality peptide deductions.
Dates Active: August 2021 — May 2023
Organizations
National Institutes of Health (NIH)
